Rice flowering and photoperiod
Rice (Oryza sativa) is a key model organism in plant biology, having contributed to numerous fundamental discoveries in the field. Like many other plants, rice enters the reproductive phase at a precise time of year when environmental conditions are favorable for inflorescence development, fertilization, and seed production.
One of the most important environmental signals regulating this transition is day length (photoperiod), a reliable indicator of seasonal change. Our research focuses on studying the genetic networks that control rice reproductive development in response to photoperiod variations.
Rice Flowering
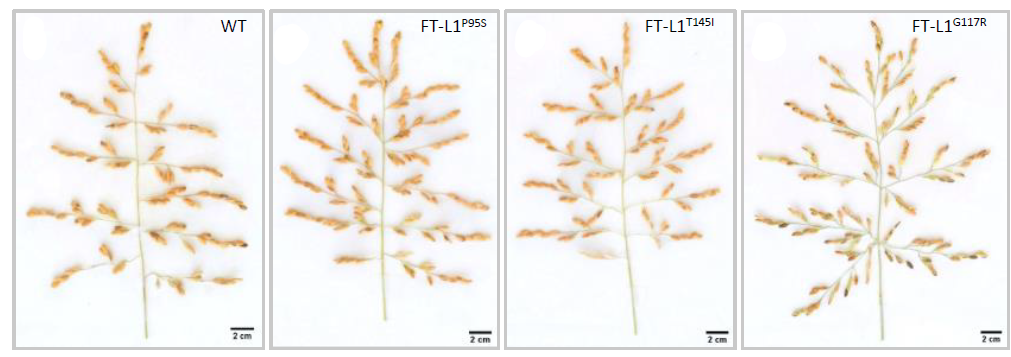
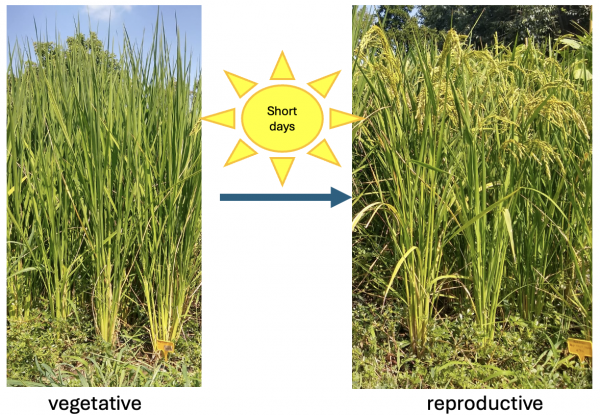
In rice, the transition to the flowering phase is triggered by day lengths shorter than a critical threshold, which can vary depending on the variety. In the image: on the right, plants in the vegetative phase, continuing to produce leaves; on the left, plants in the reproductive phase, with mature panicles clearly visible.
In model rice varieties, flowering is induced under short-day conditions, when the length of the night exceeds a critical threshold. This stimulus activates the transcription of the HEADING DATE 3a (Hd3a) and RICE FLOWERING LOCUS T 1 (RFT1) genes in the leaf phloem. The proteins encoded by these genes, known as florigens, act as systemic signals: they travel through the vascular tissues to the shoot apical meristem, where they trigger the transition from vegetative growth to inflorescence formation.
Regulation of Flowering in the Leaf
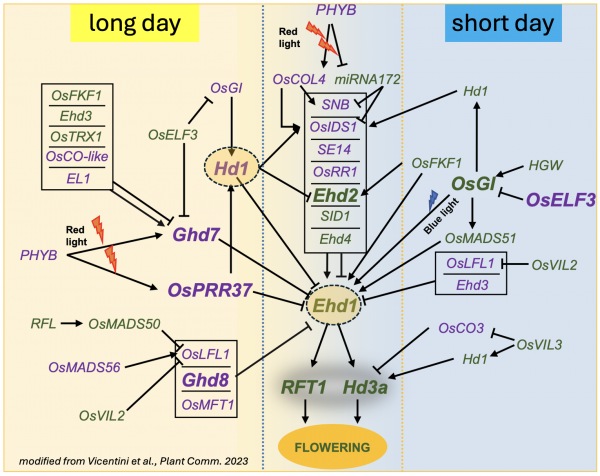
The gene networks that regulate the transcription of the florigens Hd3a and RFT1 in the leaves are complex and well characterized. Their activity is tightly controlled by the circadian clock and modulated by photoreceptors that detect light wavelength, enabling the plant to synchronize flowering with environmental conditions.
The gene networks active in the leaves are essential for establishing the spatial and temporal expression patterns of the Hd3a and RFT1 genes. Several molecular regulators—including transcription factors, kinases, and photoreceptors—translate environmental signals into responses that promote flowering. Among these, the main regulators are HEADING DATE 1 (Hd1) and EARLY HEADING DATE 1 (Ehd1), two transcription factors that activate florigen expression under short-day inductive conditions.
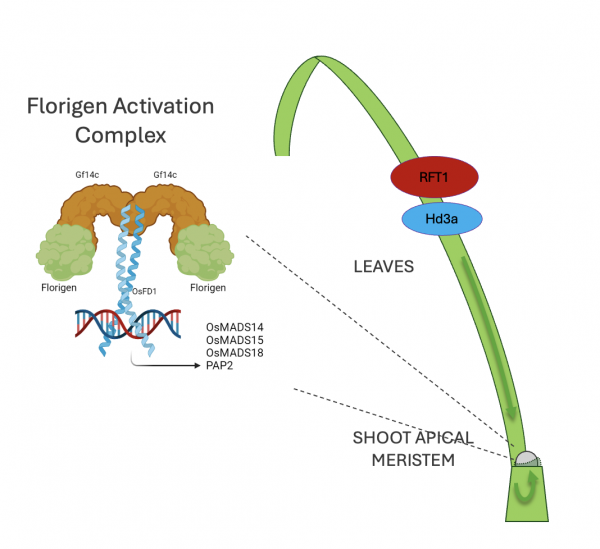
The Action of Florigens in the Meristem
When the florigens Hd3a and RFT1 reach the shoot apical meristem (SAM), they enter the nuclei of cells and form transcriptional complexes that regulate the expression of numerous target genes. Among these, several MADS-box family genes stand out as essential for triggering the transition of the meristem from a vegetative to a reproductive state.
In the shoot apical meristem, the arrival of the florigens Hd3a and RFT1 activates molecular networks that initiate inflorescence formation (see Panicle Development) and stem elongation (see Stem Elongation). The activity of the florigens depends on the formation of the Florigen Activation Complex (FAC), a heterohexamer composed of the florigens, 14-3-3 proteins, and bZIP family transcription factors. This complex binds to DNA, determining the specificity of gene activation and guiding the transition to reproductive development.
See also